How to Determine Which Enzymes to Use to Cut Plasmid
The copy number of the. Im a little confused as to how to figure out the formula needed to figure this out.

Ap Biology Restriction Enzyme Digests On Circular Plasmids Youtube
This means its important to check whether plasmid and selected restriction enzymes are.
. The tube with EcoR I enzyme has an E with a black dot in top and the tube with the BamH I. Type III cut DNA near. The enzymes and buffer are stored in the freezer.
Type II cut DNA within or close to the recognition sequence. Consult the Promega enzyme buffer chart on the last. DNA genomic is used for PCR amplicon is digested by restriction enzyme.
This is greatly facilitated by the use of specific restriction endonucleases. For a full list of REs with recognition sites within the DNA molecule select Custom. Restriction digest could possibly be used to identify the dimer even if you have an enzyme that will produce a single cut in the monomer plasmid.
Each enzyme recognizes one or a few target sequences and cuts DNA at or near those sequences. Then depending on your purpose you. Ylp has low transformation frequency 1-10 transformantsµg DNA which can be enhanced by linearizing the plasmid using restriction enzymes.
Restriction enzymes are DNA-cutting enzymes. Answer 1 of 3. Select restriction enzymes to digest your plasmid.
To determine which restriction enzymes will cut your DNA sequence and where they will cut use a sequence analysis. All have the same mass in basepairs but are in different positions because their differing shape causes them to move. Make sure you use excellent aseptic technique and change tips every time.
Each restriction enzyme recognizes and can attach to a certain sequence on DNA called a restriction site. Generally Type I enzymes cut DNA at locations distant to the recognition sequence. If you make a diagram you will find that a.
This is important because it ensures that the fragments end in a particular base AGC or T. Tyler Ford February 18 2016. I have a plasmid that is 73Kb and am told that it is cut with an enzyme of 4bp.
Many restriction enzymes make. Otherwise the electrophoretic gels could not give any meaningful sequence. One enzyme unit is defined as the amount of enzyme needed to completely digest one microgram of linear double-stranded DNA in one hour at the appropriate temperatureTo cut covalently.
When cloning by restriction digest and ligation you use restriction enzymes to cut open a plasmid backbone and insert a linear fragment of DNA insert that. Search by the number of cuts 1 find. Try choosing unique enzymes.
You can think of restriction enzymes as little molecular scissors that. It depends on what you intend to do with the plasmid but in any case you must know the exact nucleotide sequence of it and a map of its genes. Open the Digests button navigate to New Digest and specify NEB and Double Cutters in the settings.
Open and switch the view to Linear Map. Set up digest by adding ingredients in the following order. - 10 µl of 10x.
Enzymes that only cut once allow you to more easily and accurately visualize the full size of your construct. The tube with the buffer has a black top. DNA genomic is digested by restriction enzyme then an oligo-targeter is added for detection.
The ideal plasmid vectors have high copy numbers inside the cell. Its OK to set this up at room temp. On the default Graphical View page you can select 1 cutters 2 cutters 3 cutters or List 0 cutters.
If you are going to use only one restriction enzyme or enzymes that have compatible overhangs or no. Gel bands of cut left and uncut right plasmids. Buffers used in double digestions must be compatible ie both restriction enzymes should cut when combined with a particular buffer.
Consider buffer and temperature.

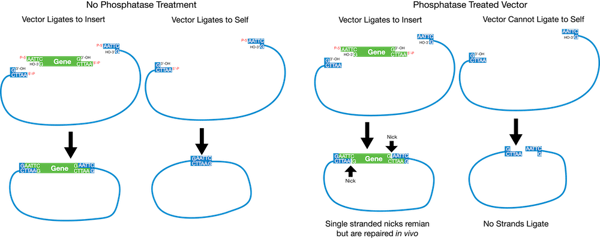
Plasmids 101 Restriction Cloning
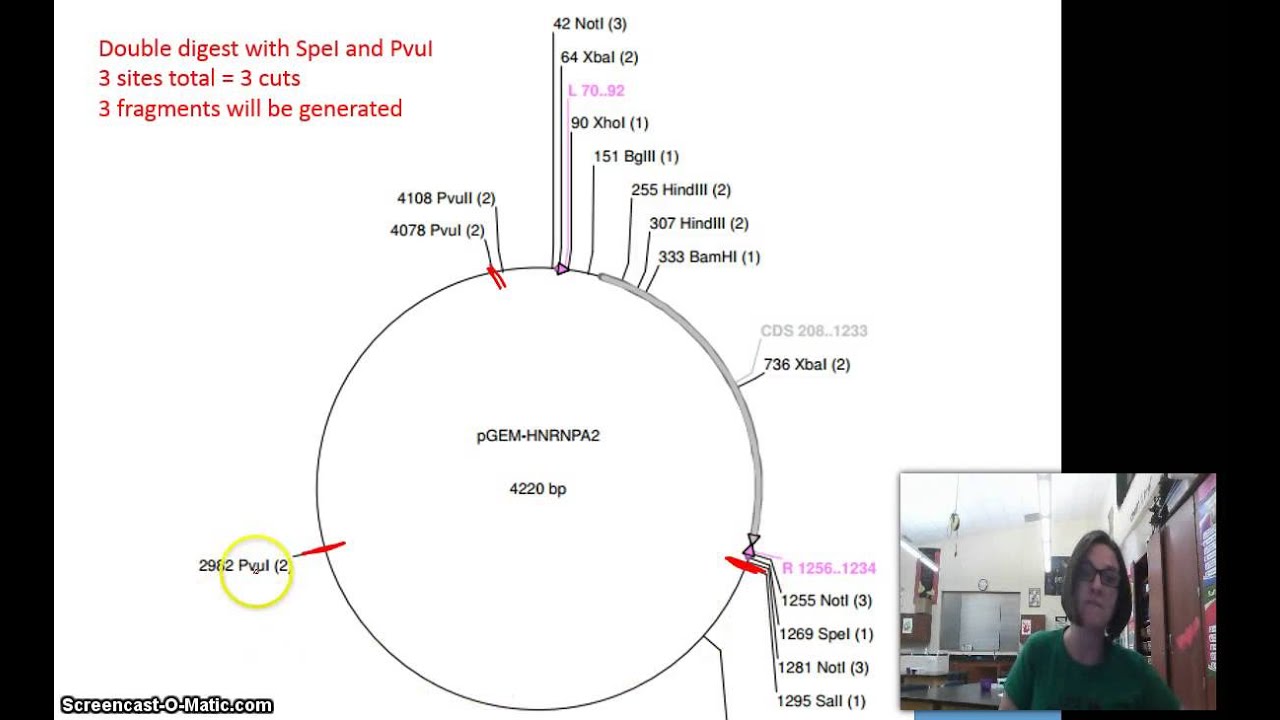
How To Read A Vector Map For A Restriction Digest Youtube
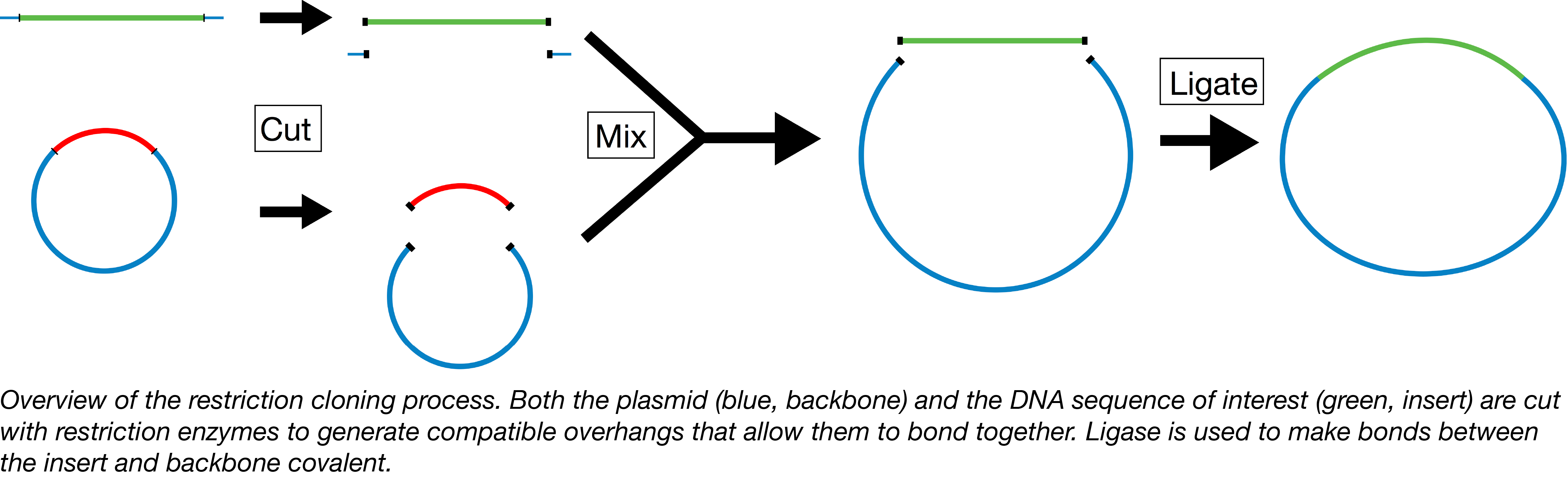
Plasmids 101 Restriction Cloning
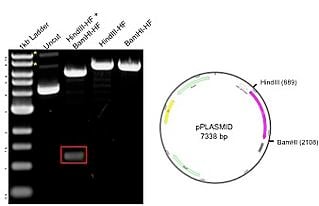
Plasmids 101 How To Verify Your Plasmid Using A Restriction Digest Analysis
No comments for "How to Determine Which Enzymes to Use to Cut Plasmid"
Post a Comment